Lab 3: NLME AND PKPD Modelling¶
This is the third computer exercise in the course TMBT42 - Systems biology, Digital Twins and AI. In this computer exercise we will look at two concepts: Non-linear mixed effect modelling and pharmacokinetic/pharmacodynamic (PKPD) modelling.
Throughout the lab you will encounter different "admonitions", which as a type of text box. A short legend of the different kinds of admonitions used can be found in the box below:
Different admonitions used (click me)
Background and Introduction
Useful information
Guiding for implementation
Here you are tasked to do something
Reflective questions
This computer exercise in developed for MATLAB, however, you are free to implement it in any environment of your choosing. Although, please note that the practical instructions below will focus on implementation in MATLAB.
To start with here are some instructions for what you'll need to install before starting with the lab. Note that if you have done the previous modules in the TMBT42 course you will likely already meet many of these requirements.
Before you start with this computer exercise, please ensure that you have the following :
- MATLAB running on your computer (or a computer in the computer hall)
- A C-compiler installed in MATLAB
-
Downloaded and installed IQM tools in MATLAB
-
Suitable toolbox packages installed. You will need tools for optimisation algorithms, statistical testing, and random sampling. In MATLAB you'll want the following toolboxes installed:
- "Global optimization Toolbox"
- "Statistics and Machine Learning Toolbox"
All files needed in this example are located in the folder NLME_PKPD inside the Lab3 files.
Copy these files to an appropriate directory.
As stated above, you are free to use any software environment however, you will need to redo the pre-made scripts and find compatible tools yourself.
How to pass (Click to expand)
At the very bottom of this page you will find a collapsible box with questions. To pass this lab you provide satisfying answers to the questions in this final box in your report. Throughout this lab you encounter green boxes marked with the word "Task", these boxes contain the main tasks that should be performed to complete the lab. There are five tasks in total, and for each task there are questions (found at the end of this page) that should be awnserd in the report. The questions can be answered in a point-by-point format but each answer should be comprehensive and adequate motivations and explications should be provided when necessary. Please include figures in your answers where it is applicable.
If everything is in place, we are now ready to start.
Introduction¶
In this exercise we will use the blood-pressure model from the previous exercise. We will first apply non-linear mixed effect (NLME) parameter estimation to fit the model to data from several indviduals simultaneously. We will then create PK model for the blood pressure medication Valsartan, where we will also use NLME to fit data for indviduals in our cohort. We will then combine the blood-pressure model and the PK model into our own PKPD model. Using this model we will then try to find an optimal dosage of valsartan to lower the blood-pressure by 10% for each individual in our cohort.
For a reminder and some context here is the description of the blood pressure model.
Model
The model used in this exercise is very simple and consists of two ordinary differential equations (ODEs) with a total of 6 parameters. The two states described by these ODEs are the SBP and DPB respectively.
The parameters \(k_{1,SBP}\) and \(k_{1,DBP}\) represents a combination of factors that are not significantly altered by the ageing process. These are typically inherenfactors such as vascular tone or cardiac function.
The parameters \(k_{2,SBP}\) and \(k_{2,DBP}\) represents a combination of factors that are directly connected to the ageing. These could be factors such as increasing arterial stiffness, reduced arterial compliance, and altered vascular function, all of which can contribute to the observed age-related change in blood pressure.
The parameters \(b_{SBP}\) and \(b_{DBP}\) represents the influence of factors that contribute to a person's baseline blood pressure level. These factors may includgenetic predispositions, lifestyle choices, environmental influences, and chronic health conditions that impact blood pressure regulation.
\(SBP_0\) and \(DBP_0\) are the initial values for the two states respectively.
Lastly, the MAP is calculated as the difference between the SBP and the DBP, divided by 3 and added to the DBP.
Exercise¶
Let's get started! Right click on the MATLAB application tilted 'lab3.mlapp' and press run.
Important to note is that
The first task of this lab will be to generate a dataset describing the blood-pressure change for all individuals in a group. As it is easy and fast to measure blood-pressure, it will also be optional to include your own, baseline data in this lab (more information during the lab occasion). This step is not mandatory, just for fun!
Task 1: Generate the data set
Use the first tab 'data' in the application to generate a dataset to use in this lab. This is done by click the "Generate data" button, which then saves the dataset to your current directory.
Optional: Add new data to the cohort
Make a measurement using the blood pressure monitor and add your data of systolic (SBP) and diastolic (DBP) blood pressure into box titled "New measured data". Multiple new data can be added via switching between the tabs 'p1', 'p2'...
Generate the dataset
The generated data will be based on the previously used Fermingham dataset. Chose the size of your cohort by using the slider in the application. You can use the "Generate data" button several times.
We have now generated a dataset. Check that you have the dataset saved in your Data directory ('cohort_SBP.mat' and 'cohort_DBP.mat'). Note that for each time you press the generate button, a new dataset is generated, and overwrites the previous one.
We will now use the NLME optimization to find individual agreement for the blood pressure model to each individual in the new dataset.
Task 2: NLME
The next tab in the application "NLME" will now be used.
The first step will be to complete and fill in the gaps in the provided script 'run_nlme', check that it exist in your directory.
We will be using the MATLAB native (Statistics and Machine Learning Toolbox) nlmefitsa function. You can check out some nlmefitsa examples. This page will be useful so keep it in mind.
Setup
The first part of the script consists of setting up all variables, options etc to be able to run the nlmefitsa function. The main parts
are the following (which are shown as individual section in the script):
Setting up the model
As in the previous labs we are using the IQMtoolbox for simulating the model. The model text file is denoted "bp_age_model.txt", check that it is present in your directory. In the script the name is stored in the variable 'modelName'
Please fill in the blanks in the MATLAB script to do the following: create/compile the model into a mex-file using the IQMmakeMEXmodel function,
create a function handle for the compiled model mex-file using str2func function, and lastly extract the model parameter names and values using the IQMparameters function. There is a code outline in the 'nlme_run'. Please complete it.
Data
The next step is to format the generated data to be compiled with desired format for the nlmefitsa function. Check out the mathworks help page for nlmefitsa. Essentially, we want to create three new variables: Y, X and GROUP:
- Y is a n × 1 column vector containing the response to each observation, n is the total number of observation in the population.
- X is a n × h column vector for all observables, where h is the number of predictor variables, in our case we only have time, so the matrix size should also be n x 1. So X only contains the time point of each observation for the whole dataset. The GROUP variable should map each observation (in X) with corresponding response (in Y) and should thus be a character matrix with rows for group names, assigning each observable to an individual.
- The GROUP variable should thus be of size n × 1 where each row is a string with a name for the specific individual/group e.g., 'pat1'
We will do this by using a for-loop, which loops over all patients (using an existing data strucutre e.g., 'cohort_SBP') iteratively building the X,Y and GROUP variables. All three variables are column vector and should contain corresponding information for all patients. Code for the for-loop is provided in the script, but needs to be uncommented. Take some time to undersand the syntax. Feel free to run the script, and look at the variables created by the loop.
Options
All code is provided for this section. The x0 variable is the start guess for the parameter estimation, and uses the paramete-values defined in the model text-file. The variable xform holds information if the parameter-space should be either linear och logrithimic. The fun vaiable, is a function-handle, which points to our simulation function; f_sim. Lastly, the 'options' variable holds som general settings for the nlme-fitsa function. Please uncomment the code.
We will now run the nlme optimization. The syntax for two different calls to nlme-fitsa function have been provided. These have slightly different settings. Try to undertstand each input in the function-call. Start with uncommenting the first one, aswell as the syntax for saving the optimization results (found at buttom of the script).
First we will run the optimization assuming some correlations among the random effects; Cov0 = ones(length(x0)) (the covariance pattern). Click on the "click to run!" button in the nlme tab of application (or run your script normally). The nlme optimization should now be running and printing in the command window. The columns correspond to iter, RMSE, B. Try to understand what is being printed, what are iter, RMSE, B? The results will be save in the NLME_FIT.m file in your directory.
What are the results of the optimization? In the nlme tab you can easily print each component, PHI, PSI and b to the command window using the small menu. You can also just look in the NLME_FIT structure. What are the fixed and random effect? Spend some time to understanding each component (i.e., check nlmefitsa link), this will be of benefit later.
We now want to make PSI a bit more easily understandable by converting it to the correlation matrix RHO. Easily done via corrcov.Please write the MATLAB synthax for RHO in the small text box (in the nlme tab of the application) and click the 'Evaluate' button. Please save the figure by clicking the 'Save figure' button.
Now comment (by putting '%' before the function-call) or remove the first call to nlme-fitsa.
We will now do the same thing again but assuming no correlation. Uncomment the secound function-call, and run the script again.
Print RHO again and save the new figure.
Alright! We now have found the fixed and random effect parameters describing all individuals in our data set. We should now look how the simulation fit to data!
Task 3: Model agreement using NLME result
We can now move on to the 'NLME fit' tab in the application. Alternatively, create your own new separate script for plotting.
Simulating the fixed effect
We will start by simulating the model with only the fixed effects. In the application the name for the parameter vector for model simulation in THETA. In the first box, we will let Theta only account for the fixed effects. Please write the following in the first box:
Theta = exp(PHI')
Keep in mind that the parameters where stored in log-space, and thus need to be transformed before the simulation.
Click the 'evaluate' button, to run the model simulation. The blue dotted line in the figures are the model simulated using only fixed effects.
What are the bar plots to the right?
Simulating the fixed effect and random effect
We now want account for both fixed effect PHI and random effect b. How should this be done? The goal is to create a matrix where each row represent the model parameters for one individual, that accounts for both the fixed effect and the individual random effects. Please write the following in the second box:
Theta = exp(repmat(PHI', num, 1)+b')
Take a moment to understand the syntax. The variable 'num' is the number of indviduals in our data.
Click the 'evaluate' button, to run the model simulation. We should now have simulations for each individual. What did change in the bar plots to the right?
Save the figures by clicking the 'Save figures' button.
Great! We have now done a first basic nlme fitting!
We will now move on the phamcokinetic/phamcodynamic (PK/PD) part of the exercise. In short, a PK model describes how the body metabolizes the drug, and a PD model describes how the drug effect the body. Would you characterize the model we have used up to this point as a PK, PD or PK/PD or model?
The drug we will be looking at in this exercise is Valsartan. Valsartan is a angiotensin II receptor type 1 (AT1) antagonists. The drug is rapidly absorbed after oral administration, and a typical dose is 80-320 mg. Valsartan has the ability to reduce blood pressure, and is thus often used to treat patient with high blood pressure.
The mechanism of action is by blocking the actions of angiotensin II, which is a peptide hormone responsible for vasoconstriction. Vasoconstriction is the narrowing (constriction) of blood vessels by small muscles in their walls. Valsartan blocks this action, resulting in lowering of blood-preesure.
The next step will now be to create a simple PK model for Valsartan, using generated data for our own cohort!
Task 4: Pharmacokinetic model for Valsartan
Lets now move to the tab named 'PK' in the MATLAB application.
Generate Valsartan data for the cohort
Click the 'Generate data' button. This will generate a dataset describing a single oral dose of 160 mg Valsartan and the corresponding plasma concentration over 24 hours for each individual.
Creating the PK model
In the text box too the right we have an unfinished version of the PK model. We will use a two compartment first order elimination model. The interaction graph for this model is shown below.
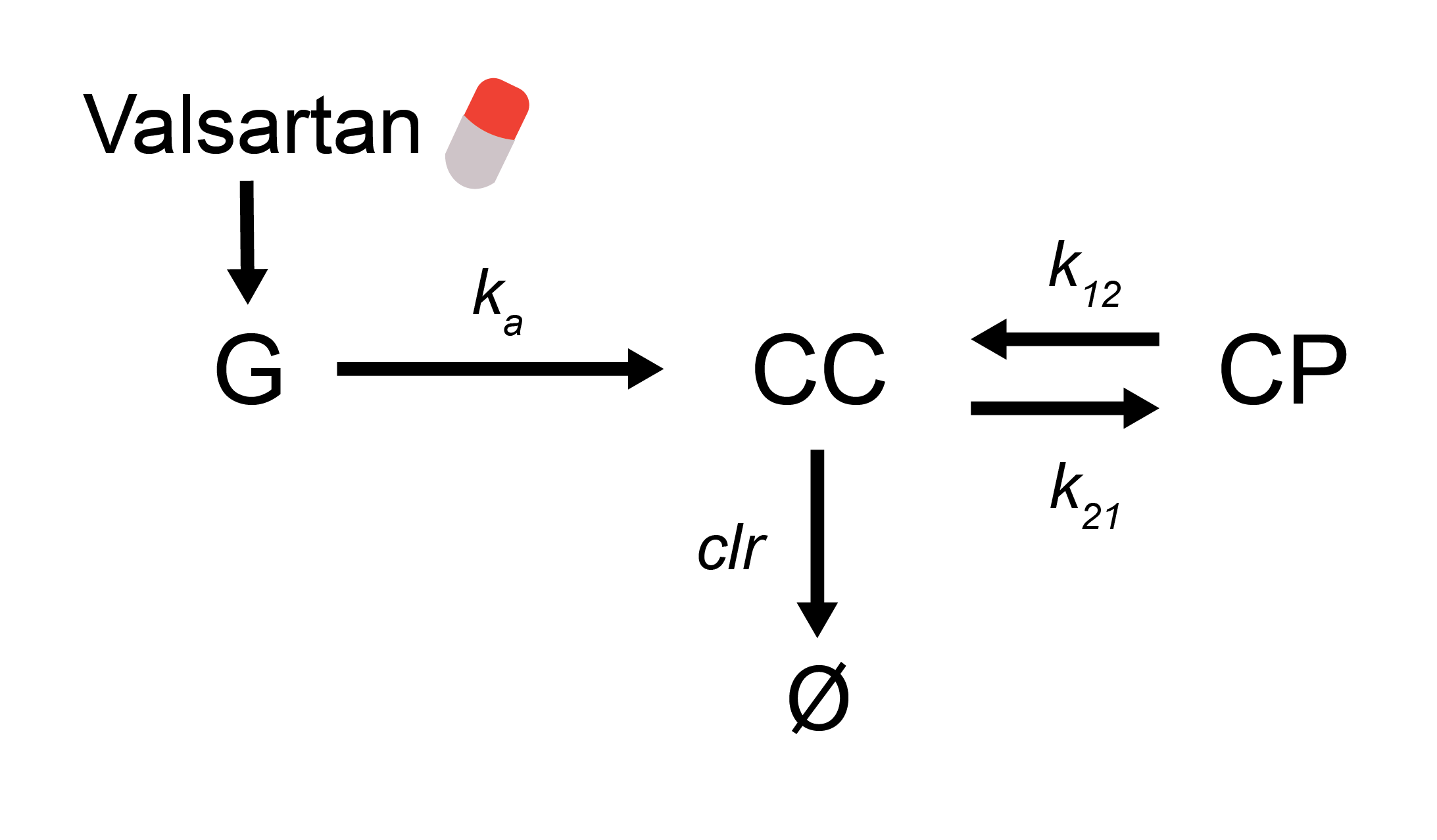
Use the interaction graph to complete the model in the text window. The parameters values you give in the model can be arbitrary, but the initial conditions all need to be zero. Why? Please, do not change the names of any model states, or the name of the model. Here, we define the dose of Valsartan in the Dose parameter (do not change this name), and give the dose in the events already defined.
Click the 'create model' button. This will save the model into a text-file called 'PK.txt' in your current directory.
Now click on the run nlme button. Can your model fit to the Valsartan data? If so, great work! Now, save the figures and lets continue! To note, getting a CVODE Error: CV_ERR_FAILURE might be result of an error in the model formulation. Please check it.
The answer to the question about how to characterize the blood pressure model (PK, PD or PK/PD), is that the model is neither. However, the model could be combined with a PK model to function AS PK/PD model. Let's do that!
Task 5: Building a PK/PD model
We can now move on to the PKPD tab in the MATLAB application.
Note that there is a difference in the time-scale of the blood pressure model and our new Valsartan PK model. The blood pressure is simulated over years and the PK model simulates plasma Valsartan over hours. Click the 'Simulate 24 hours' button (left side of the interface). We here simulate the blood pressure model for 24 hours (instead of 70 years). How do you think the time vector for for this simulation looks like? Here, we see that the blood pressure is unchanged during the 24 hours (which is a crude approximation, but will work in this exercise).
With this important technicality explained, lets go on with building the PKPD model.
Building the PKPD model
The textbox contains the model equation for the blood pressure model, which will function as the PD part of the model, as we want to simulate Valsartans effect on blood pressure. Now, combined the two models! Note, do not change the order, position or names of the last two parameters ('Dose' and 'number_daily_doses').
So, Valsartan is still not asserting any effect on the blood pressure. Thus, we now need to create a variable describing the effect of Valsartan on blood pressure. In the textbox, a model variable called VE (Valsartan Effect) is already created. Please make it so that VE has a saturated dependency on the concentration of Valsartan in plasma (CC). Suitable parameters are Imax = 35, and IC50 = 0.25. What unit should these parameters have?
We have created the effect variable. However, we still need to add VE so it changes blood pressure. We will do this very simply, partly because of the nature of our PD model. Please add VE as a negative term in the SBP derivative, this term should also dependent on the current SBP value, e.g., VE*SBP. What should happen to blood pressure if we introduce Valsartan?
Click the 'Simulate PKPD' button to simulate the PK/PD model! What does each figure represent? Does it looks like the varible VE have similar time-dependcy as the plasma concentration of Valsartan?
Do we see a change in blood pressure during these 24 hours?
Now, save the figures, by clicking the 'save figures' button and lets continue!
We have now created our own PK/PD model for Valsartan's effect on blood pressure. Let's now jump to the 'multiple dose' tab. Here we can make model predictions of different Valsartan dosing schemes; with different doses, different number of doses per day, and for different amount times.
Is it possible to find a dosing scheme that would lower the systolic blood pressure by exactly 10% for all patients by the end of 7 days? Play around and see if you can do it! The answer is probably not!
Each individual in our cohort is different, and thus the dose fulfilling this criteria is probably different for each individual. However, we have the differences quantified in our nlme parameters. So lets see if we could calculate an optimal dose for fulfilling this criteria for each individual.
Task 6: Optimal Valsartan dose
Let's move to the optimal dose tab in the MATLAB application.
This will be an optimization problem. However we will not use nlme. In this case it easier to use a 'regular' optimization algorithm function e.g., particleswarm. We will thus need to create a cost-function. We have provided a new cost-function denoted 'cost_PKPD_optimal_dose' and should be in your directory. Open this file.
Completing the cost-function
Here, we have provided syntax for simulating the model for each partient using their specific parameters. Please take sometime to understand the syntax. Essentially, we are using a for-loop to simulate each patient. We should now also add syntax for adding the criteria of lowering blood pressure 10%.
To satart we should calculate the relative change in blood-pressure (SBP) after the intervention for each patient.
First, define the variable 'change' as the relative change in blood-pressure. The strucure sim, is the result from the model simulation (from one patient). The values for blood-pressure (SBP) is thus saved as : sim.statevalues(end,ismember(sim.states,'SBP')). Use this infomration to define the 'change' variable.
We should now add the citeria of lowering the blood-pressure (SBP) by 10%, ergo we want change to be equal to 10. One way of solving this is to calcualte the residual : (change - 10)^2
We want to save this calculation for each pateint. This could be done by letting a varible grow for each loop iteration e.g.,
r(i,:) = (change -10)^2 ;
The last step is to define the function output, the 'cost' variable. Please uncomment the 'cost' variable , and define it.
When working we should be able to find estimated for the optimal dosing scheme for each patient.
When you have first version of the function. Press the 'Run optimization' button. This might take a few minutes, so look through the questions to be answered in the report (found at the of this document).
Now we are done!
Check so that you have saved all figures in the figures folder in your directory.
Please awnser the following questions in your report.
Summary - Questions for lab 3 - NLME and PK/PD:¶
In your report, you should present answers to the following questions
Task 1 - Dataset
- Please describe the dataset brifley. What are some fundamental criteria for dataset to be able to run nlme paramter estimation?
Task 2 - NLME
- What does mixed effect entail? What is an NLME model? Please describe brifley, include equations.
- We used two different settings for running nlme. This resulted in different RHO matrix. Please explain the difference.
- Please explain the result from the nlme optimization. Brifley, describe each variable, PHI, PSI and b.
Task 3 - NLME result
- Please describe the figures. What is being plotted? Explain the second formulation of Theta.
- In which model parameter do we see the largest variance? Why do we see this? Please discuss.
Task 4 - PK model
- We are missing some fundamental parts in the PK model, which are usually included. What might these be? Please discuss.
Task 5 - PKPD
- What units are IC50 and Imax in? Theoretically, what would happen if IC50 increased or decreased?
- Could there be another way to implement the effect VE into the SBP derivative? Please discuss.
Overall
- What are som core principals behind NLME. - When is NLME useful, and when is it not?